SBGrid Consortium homepage

Structural biology software, anywhere you work - read more. Supported by Members, with additional funds from NIH, NSF, Helmsley and AWS. Cite the SBGrid Acta Crystallographica paper.

Read our January newsletter
Latest upgrades from SBGrid, including latest software updates, new members and special events. Published montly by SBGrid.

SBGrid Webinar Series
Join us for our Software Webinar Series - Tuesdays at 12pm ET - to hear from software developers about what's new in structural biology. Community members connect via Zoom. Subscribe …

Capsules Technology
We have recently published a manuscript describing our Capsules technology used to deploy SBGrid and BioGrids software collections. Read more in Acta Cryst Section D, special 75th ICUR anniversary issue, …

Connecting SBGrid with Instruct-ERIC
SBGrid is pleased to announce that it has established a partnership with Instruct-ERIC, based in Oxford, UK. Instruct-ERIC is a pan-European distributed research infrastructure with a hub in Oxford, UK …

Reshaping Membranes
Cryo-electron microscopy has been a boon for the study of membrane proteins. No crystallization needed, and the ability to visualize dynamic regions of the protein to boot. But one aspect …
-
Report Software Bug
Software fixes, depending on priority take 1-14 business days to complete.
-
Request New Software
New software packages are added on a monthly basis, or more often when requested.
-
Other Support
Any other requests or comments can be submitted here.
-
Grant Support
A statement for your resources section, support letters, and how to cite SBGrid.
Epitope-spanning antigenic variation reprograms immunodominance and broadens immunity in sequential influenza vaccination.
Computational approaches for RNA structure prediction and design.
PrePPI - Structure-based prediction of protein-protein interactomes and networks.
-
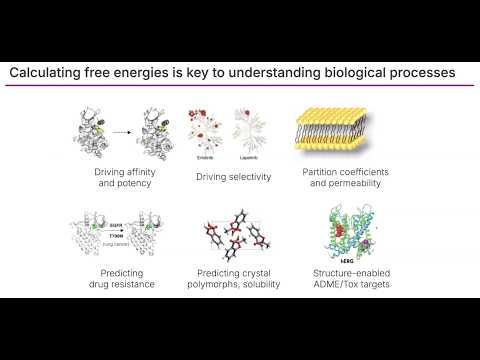
Open Free Energy - An Open Source Ecosystem for Calculating Free Energies
-
CryoDRGN-AI: Neural ab initio reconstruction of challenging cryo-EM and cryo-ET datasets
-
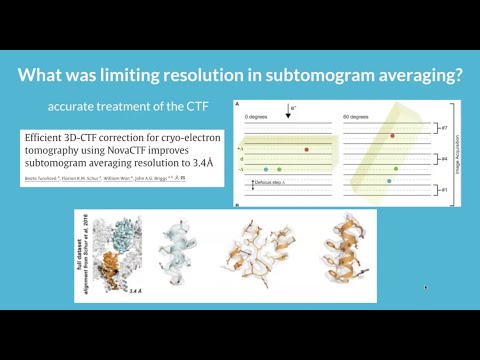
Warp
-
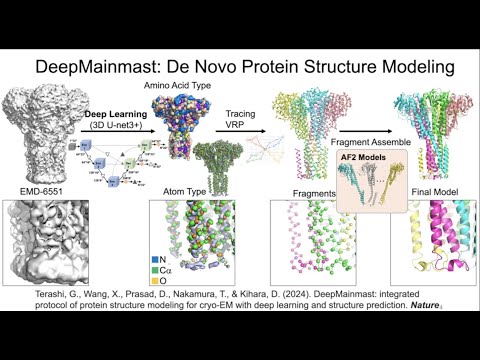
DeepMainmast and DAQ
-

MemBrain v2
-

AreTomoLive - automated cryo-electron tomographic alignment, reconstruction, and contrast enhancemen
-

Phenix/AQuaRef
-
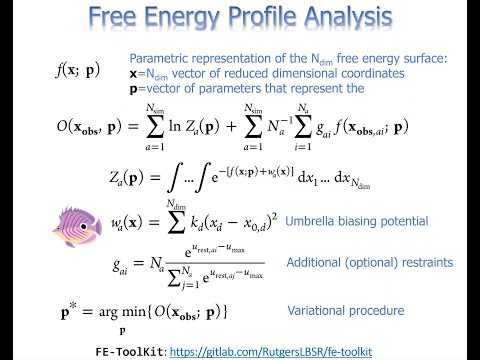
Free energy methods in Amber
-
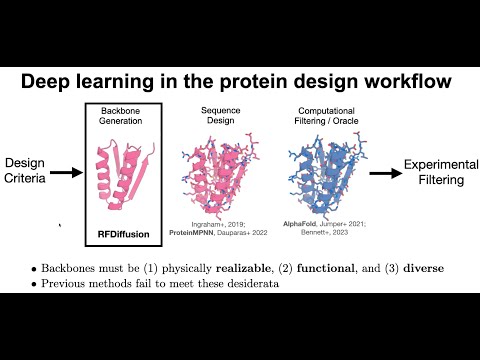
De novo design of protein structure and function with RFdiffusion.
-
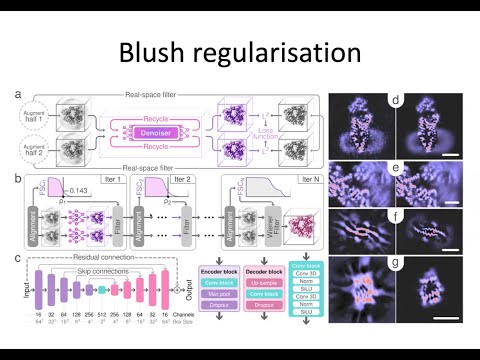
RELION-5.0: Recent developments






