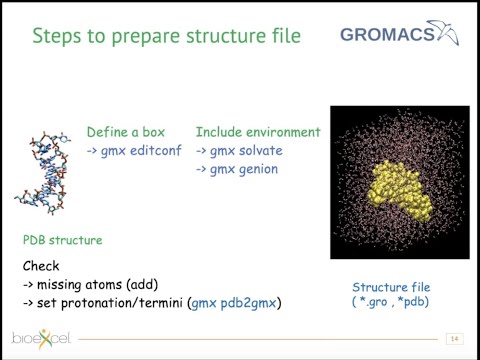
Supported Applications
GROMACS
-
Description
a versatile package that performs molecular dynamics of proteins, lipids and nucleic acids.
-
Usage
To list all executables provided by GROMACS, run:$ sbgrid-list gromacs -
Usage Notes
SBGrid Usage Info
GROMACS is GPU-accelerated using Nvidia CUDA on Linux. For general information on running GPU accelerated applications from SBGrid, please see here : https://sbgrid.org/wiki/gpu
CUDA builds of GROMACS are designated by a '_cu' suffix in the version of the application. For example, 2020.2_cu9.2.88 is linked against CUDA v9.2 libraries. All builds can be run on 'consumer-grade' GPU hardware.
GROMACS builds which contain _plumed are compiles with PLUMED, https://www.plumed.org .
2020.2_sse2_cu9.0 is compiled for older hardware (pre-intel Haswell CPUs, circa 2013)
GROMACS and MPI versions
GROMACS uses MPI and OpenMP for parallel processing. MPI is managed by the application and should not need to be explicity set in the shell. Please let us know if you have MPI problems or questions - bugs@sbgrid.org.
Recent Changes
20200703 : Add 2020.2_sse2_cu9.0 compatibility version
Deprecated versions
The following verions are deprecated and will be removed in a future SBGrid update, approximately one month from the date listed.
20200706: 2019.1 2019.1_cu9.0 2019.5_cu9.2.88 2020.1 2020.1_cu9.2.88
-
Installation
Use the following command to install this title with the CLI client:$ sbgrid-cli install gromacsAvailable operating systems: Linux 64, OS X INTEL -
Primary Citation*
M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess, and E. Lindahl. 2015. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1: 19-25.
(Note: Volume is actually 1-2.)-
*Full citation information available through
-
-
Webinars
-
Keywords
-
Default Versions
Linux 64: 2025.3_cu12.2 (305.0 MB)
OS X INTEL: 2025.3 (1.2 GB) -
Other Versions
Linux 64:
2019.4_cu9.2.88_plumed (1.3 GB) , 2019.6_nocona (391.8 MB) , 2020.6_cu10.2 (641.3 MB) , 2020.6_nocona (427.4 MB) , 2021.1_cu10.2 (801.1 MB) , 2021.2_cu10.2 (801.1 MB) , 2021.3_cu10.2 (866.7 MB) , 2021.4_cu11.5.2 (754.4 MB) , 2021.5_cu11.5.2 (1.0 GB) , 2022.1_cu11.5.2 (849.1 MB) , 2022.2_cu11.5.2 (849.1 MB) , 2022.4_cu11.5.2 (885.3 MB) , 2023.3_cu11.8 (1.1 GB) , 2023_cu11.5.2 (1.4 GB) , 2024.1_cu11.5.2 (1.1 GB) , 2024.2_cu11.5.2 (1.1 GB) , 2024.2_cu12.2 (1.1 GB) , 2024-rc_cu11.5.2 (1.1 GB) , 2025.1_cu11.5.2 (826.5 MB) , 2025.2_cu12.1 (305.2 MB) -
OS X INTEL:
2019.1 (53.1 MB) , 2021.1 (191.4 MB) , 2021.2 (191.4 MB) , 2021.3 (191.4 MB) , 2022 (85.3 MB) , 2022.1 (114.7 MB) , 2022.2 (121.0 MB) , 2023 (110.0 MB) , 2024.1 (1.9 GB) , 2024.1_arm (1.8 GB) , 2024.2 (3.4 GB) , 2024.2_arm (3.0 GB) , 2024-rc (68.8 MB) , 2024-rc_arm (1.6 GB) , 2025.2 (1.5 GB) , 2025.2_arm (2.1 GB) , 2025.3_arm (964.3 MB)
Developers
Berk Hess, David Van der Spoel, Erik Lindahl