Spiraling into focus
Carsten Sachse
Forschungszentrum Jülich
Published March 29, 2020
Some things take time and patience. Like generating higher atomic resolution structures from cryo-electron microscopy. Or reconstructing a protein’s hidden coils in atomic models. Or mastering the skills and teamwork in field hockey.
Carsten Sachse never played field hockey at the level of Germany’s world-dominating national team, but years of the sport in his youth helped shape his personality and work ethic. There’s the mix of a daily routine interspersed with intense match-ups. Rewards come from persistence and steady strategic steps toward a longer-term goal.
“To look at a topic from many different angles takes time,” Sachse says. “It feels like a marathon until you publish the paper. Then it’s certainly gratifying.”
The Sachse lab specializes in the spiraling parts of the protein universe known as helices. They probe the atomic twists in molecular structures on two distinct playing fields—image processing methods and wet bench biology.
Among SBGrid members, Sachse might be best known for a software suite called SPRING (short for Single Particle Reconstruction from Images of kNown Geometry), a suite of 20 programs able to diagnose and analyze the repetitive helical units.
On the biological front, the Sachse group probes the machinery of autophagy, a continuous cellular disposal and recycling system that rids the cell of large undesirable objects, such as damaged organelles, protein aggregates, and invading viruses and bacteria. Problems with the autophagy quality control system in cells are linked to cancers, infections, and neurodegeneration. Autophagy also kicks in during starvation and other stress to redeploy cellular building blocks.
The autophagy machinery deploys more than 40 molecules in a two-part process: engulf cellular debris and deliver them to the lysosome sacs, where the debris is broken down into reusable components.
Scientists first thought the machine moved through the cell, like the Pac-Man computer game, chomping away at all trash. Now, researchers know the process is often highly selective.
Sachse has zoomed in on one aspect of the recycling choreography, a receptor component called p62. Specifically, his team found that the p62 protein confronts the autophagy challenge with growth worthy of a comics superhero, assembling itself into helical filaments to form a large enough structure to tackle oversized debris.
“The cargo is huge on a molecular scale,” Sachse says. “The cell needs to find a way to recognize and capture larger cargo. We believe the filament organization is a way to do that.”
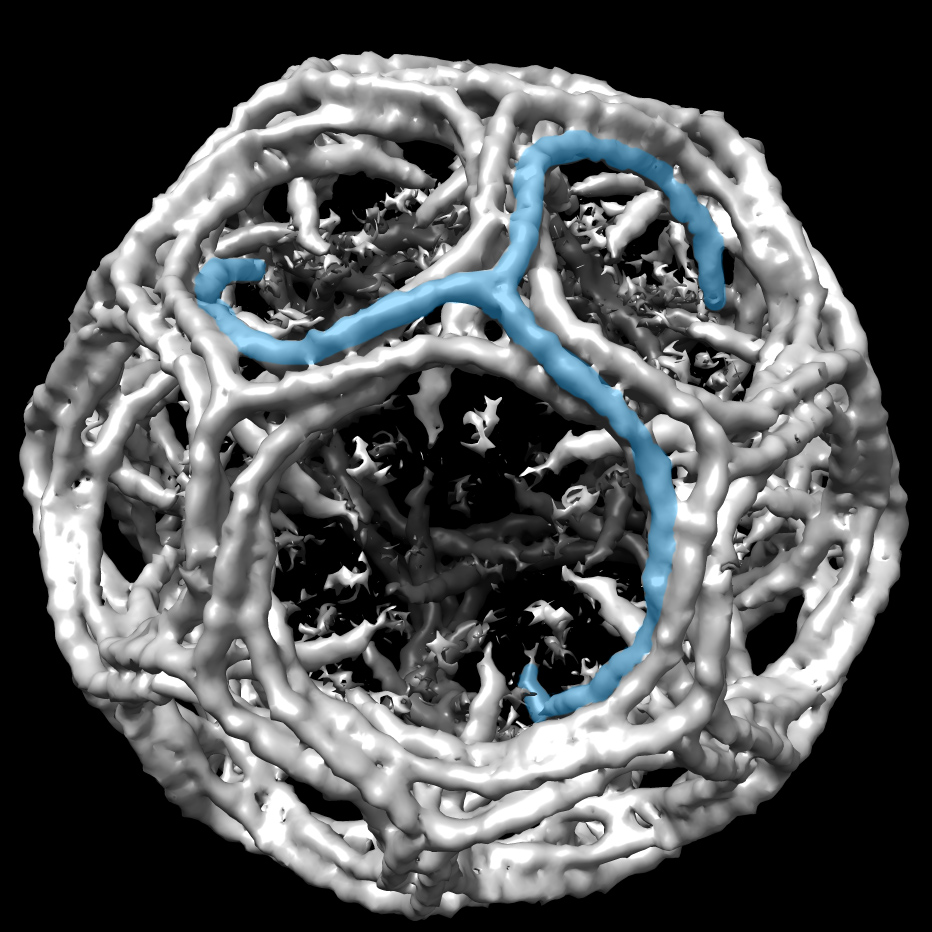
Sachse’s group has published several papers on p62, starting in 2015 with the ubiquitous pseudo-atomic resolution (10 angstroms) so many cryo-EM researchers faced at the time. The latest study, published January 2020 in Nature Communications, reported the first images of the spiraling chains at 3 angstrom atomic resolution, (using the helical reconstruction methods his lab developed).

The January 23rd, 2020, *Nature Communications* cover featured the helical assembly structure of autophagy receptor p62/SQSTM1 (blue) at atomic resolution, with the isolated p62 monomer unit (gold) that forms chains to bridge the cell waste disposal machinery to the lysosomal bins.
As an undergraduate at Friedrich-Schiller University Jena in Germany, Sachse was drawn to biochemistry as a new frontier. “It’s amazing what nature is able to do, he says. “It’s the ultimate teacher.”
His doctoral project focused on the structure of amyloid fibrils, coiled proteins that clump together in organs and are associated with progressive disease, such as in the brains of people with Alzheimer’s. It was a collaboration between the German lab of Marcus Fändrich and the U.S. lab of Nikolaus Grigorieff. Sachse soon relocated to the Grigorieff lab at Brandeis University in the Boston area.
Those were early and exciting days of learning cryo-EM, and Sachse worked on new methods using structural biology’s molecular standard-bearer, tobacco mosaic virus.
Sachse continued working on amyloid structures and new methods in short postdoctoral fellowships with Fändrich and then with Roger Williams and Richard Henderson at the Molecular Research Laboratory in Cambridge, UK.
In 2010, Sachse moved to Heidelberg to become a group leader at the European Molecular Biology Laboratory (EMBL). He shifted his biological focus from problem-causing amyloids to solutions-oriented autophagy, where new helical structures were waiting to be elucidated.
As a PhD and postdoc, Sachse was expert in imaging processing, with many colleagues approaching him to collaborate in solving structures. Now, his own lab was generating samples and proteins—something that was very time consuming. “Computers always give you output,” Sachse says. “Making proteins doesn’t always result in useable output.”
In 2014, the Sachse group released SPRING, a combined Fourier-based and single-particle workflow to delineate and determine the helical structures. The software helps people looks for the smaller units that make up the helical structure, like “little beads strung on a spring,” notes Sachse. “Seeing those beads in focus is tricky. Basically, most of the time they are not comprehensible to the human eye.”
He continued to explore helical reconstruction issues that emerged with new hardware in the early days of direct electron detectors. A 2015 paper led by PhD student Simon Fromm detailing a specific type of radiation damage and validating the work-around procedure to generate a near-atomic model was highlighted in 2018 as paper of the year by the Journal of Structural Biology.
In addition to helical reconstruction methods, his group recently developed a series of cryo-EM map interpretation tools such as confidence maps and LocScale that assist in faithful atomic model building using cryo-EM densities.
EMBL terms are limited, so in 2018 Sachse moved to Forschungszentrum Jülich, a national research center tucked into the northwest corner of Germany near the Dutch border. There he runs the structural biology program at the Ernst-Ruska Center. The center mostly houses and offers large-scale infrastructure to support material science EM, nuclear magnetic resonance (NMR), and X-ray crystallography. Some pioneering optical technologies in material science EM have been developed at the center.
In this setting, Sachse aims to establish a unique cryo-EM facility for structural biology research. “We are using this impressive infrastructure to build new machines suitable for structural biology,” Sachse says. “Cryo-EM has grown a lot, but there is definitely room for improvement in imaging quality.”
Sachse would like to be able to use cryo-EM to visualize native cellular processes inside cells. “Microscopically that is a vast space to explore,” he says. “Once we’ve imaged the complexity of a cell, there will be many new biological and technical questions. New methods will need to be developed to extract yet more information, all of which will help us to better understand how cells work.”
-Carol Cruzan Morton