Designer drugs
Ho Leung Ng
Kansas State University
Published February 25, 2021
The COVID-19 pandemic has schooled scientists in unexpected ways. Ho Leung Ng, for example, was surprised to learn coronavirus infections are common and often fatal in cats.
“I’m more of a dog person,” says Ng, whose training was in chemistry and human medicine.
Until the pandemic loomed, cats and coronaviruses were far from his mind. Ng’s laboratory at Kansas State University mainly worked on designing cancer drugs using artificial intelligence and X-ray crystallography.
In early 2020, as COVID-19 spread across national borders, Ng’s lab shifted priorities. “We felt like we were in a position to possibly make a contribution to the scientific efforts,” Ng says.
He knew other like-minded people in related fields. “There was a spirit among scientists who felt like this is possibly one of the biggest medical and scientific emergencies of our careers,” he says.
Ng redeployed his group’s computational and experimental drug design skill set. He started the Open Source COVID-19 drug discovery consortium, modeled after a similar long-standing malaria collaboration he had joined for a side project in the lab.
In March 2020, the Open Source COVID-19 researchers launched their efforts on the design, synthesis and testing of potential SARS-CoV-2 small molecule drugs. Ng and the initial research collaborators evaluated their strengths and interesting questions they could answer.
They decided to focus on the main protease of SARS-CoV-2. Inside an infected cell, the viral RNA is translated into one long single protein. The virus protease cleaves the protein into active working units.
The search for a SARS-CoV-2 protease inhibitor seemed even more promising, because researchers could build on research conducted for the first SARS outbreak, which was contained in 2003.
“From a purely scientific perspective—and this is different from the medical and epidemiological aspects—SARS-CoV-2 is closely related to the first SARS virus,” Ng says. “A lot was published. There were efforts toward making a drug, but no final drug. After that epidemic died down, all the funding disappeared. We picked up where others had left off. We were not starting from scratch. That’s very important.”
The global consortium team has computationally screened thousands of previously studied drug candidates. Ng’s team experimentally validated one interesting candidate with a collaborator. Then they learned it had been originally discovered on his same campus by researchers down the street whom he had not met.
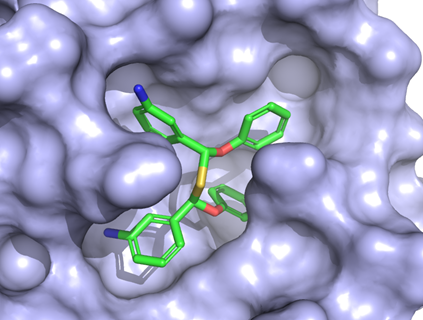
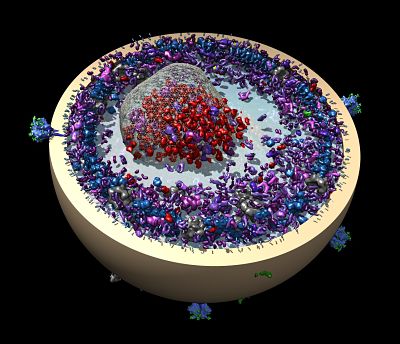
An artificial-intelligence generated drug is bound to the main protease of the SARS-CoV-2 virus. Courtesy H.Ng.
“It was developed as an experimental veterinary drug for cat coronavirus,” Ng says. “There’s a broad family of coronaviruses. A coronavirus that infects cats is relatively well-known and scary infection, usually fatal.”
Ng and his colleagues had screened for covalent inhibitors of the protease. A covalent inhibitor chemically reacts with the drug target, locking down and disabling the protease.
From a research perspective, the chemical bond requires something different from conventional computational drug screening. “Now you have to model chemical reactivity, which you normally don’t have to do with traditional methods,” he says. Ng’s group uses the Schrödinger software provided by SBGrid, but it’s a labor-intensive process.
The experimental lead molecules are tested by collaborators at the Wistar Institute. Ng's lab is working on chemical changes to improve the potential safety profile of the drug. “COVID-19 affects many people, but most people will recover,” Ng says. “It’s important that any drug we give be very safe. It can’t be more deadly than the disease.”
Pfizer has a similar anti-viral compound with some chemical modifications that has moved into phase 1 chemical trials in people, Ng says. “That feels validating,” he says, but there’s room for another drug. The Pfizer drug has to be administered intravenously, which is less accessible to people in low-income countries. Ng and his collaborators are working toward an inexpensive pill. “To attack this huge pandemic,” he says, “we’re going to need lots of different ways to take care of the virus—vaccines, drugs.”
So far, the researchers have identified 40 other molecules. They have tested them against the viral protease enzyme in lab dishes. If all goes well, cell culture testing will follow.
Synthesizing new versions of the compounds is going slow. One reason is lab disruptions when people exposed to COVID-19 must self-quarantine. Another major delay comes from lack of support. “There’s almost no funding for viral drugs,” Ng says. “A lot of us working in this field don’t understand why.”

Ng grew up in Honolulu, Hawaii. His parents had little formal education, but he was interested in science from an early age. He became fascinated with viruses, bacteria and medicine in elementary school, seeking out books on specific science topics at the library.
He discovered structural biology as an undergraduate at Harvard University in Boston. He pursued an MD/PhD at University of California, Los Angeles. After two years of medical school, he realized he wanted to focus more on research and not practice medicine. He moved to University of California, Berkeley, to postdoc in the lab of Tom Alber.
In Berkeley, he developed a form of leukemia that might have killed him just a few years earlier. Instead, his diagnosis coincided with a new generation of rationally targeted therapies developed by colleagues at Berkeley and Harvard. “I was in the right place at the right time to benefit from drugs that had just come out,” he says.
Wavering between a career in academia or industry, he worked two years as a senior scientist at ConfometRx on the crystallography of G-protein coupled receptors, a company founded by future Nobel laureate Brian Kobilka of Stanford University.
Then, a job that seemed tailor-made for him opened up at University of Hawaii, where his parents still live. It was a dream opportunity to move back home. He gave a TEDx talk about drug design and his experience as both a scientist and a patient. Speaking in front of the home crowd helped cure his anxiety about speaking to large groups, an experience he now embraces in classrooms and at conferences.
In 2017, Ng moved his lab to Manhattan, Kansas. “That leads to the question I’m asked all the time: Why leave Hawaii for Kansas?” His reasons: Lower cost of living, nice family environment, and more resources for research.
In Kansas, lab members have placed high in competitions to predict how different drugs bind to protein targets. Their drug design research is split between estrogen-related breast cancer and immunomodulators that stimulate the immune system to attack cancer.
-Carol Cruzan Morton