Beyond Cool
How SBGrid grew out of X-ray crystallography and assimilated the future of structural biology
Published May 31, 2019
With the 2017 Nobel Prize, cryo-electron microscopy became briefly famous outside of the growing clutch of scientists using the techniques to capture detailed views of molecules that keep life humming along.
The prize shone a spotlight on three scientists (including SBGrid member Joaquim Frank). Beginning in the 1970s, they pioneered a trifecta of advances in sample preparation, imaging, and software. Many more scientists and software developers have built on these key advances to make cryo-EM a new force in modern structural biology.
On a smaller scale, an entirely different trio denoted the future of cryo-EM for SBGrid. In 2004, a triad of papers from collaborating laboratories in the Boston area provided an early glimpse of the bigger potential of EM for atomic resolution and helped SBGrid identify key software platforms to support the emerging new branch of structural biology. The research happened at a time when SBGrid was evolving out of a single lab and becoming a global research computing consortium.
“Those three papers represent an expansion of structural biology well beyond X-ray crystallography at a key stage where the technology became integrated into SBGrid,” said Steve Harrison, who heads the Howard Hughes lab at Harvard Medical School (HMS) where SBGrid started nearly 20 years ago. “They allowed SBGrid to become the universal software platform for structural biology that it now is.”
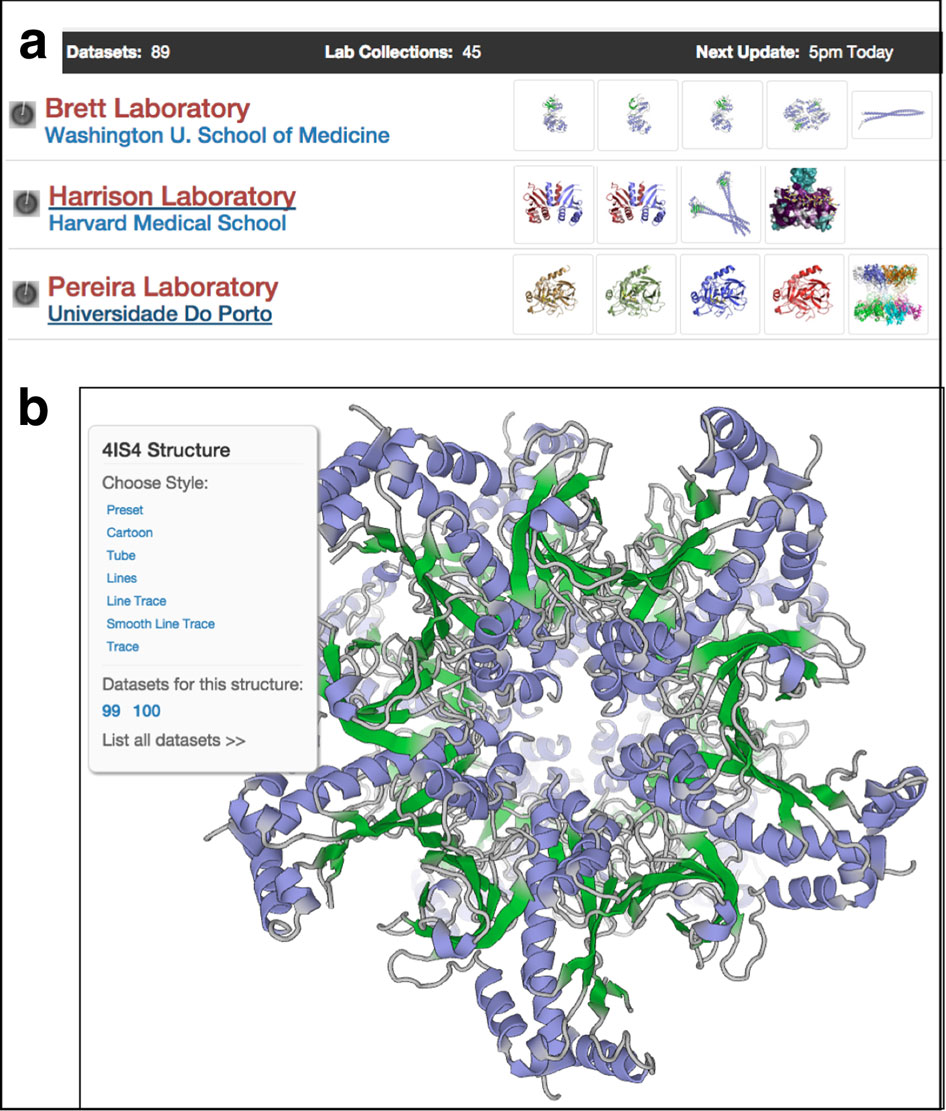
SBGrid recently took a look back at the three projects that led its way into EM. In 2004, SBGrid linked more than 100 HMS computers. The grid offered a supportive computing environment and speeded up the calculations needed to solve complex protein structures from weeks to hours.
Authored by overlapping research teams, the studies determined structures of a clathrin coat (and a coat bound to a co-factor of its dismantler auxilin), transferrin and its receptor, and aquaporin-0. The molecules and their mates were either too big, greasy or fragile to grow into the rigidly ordered rows of hard crystals needed for X-ray diffraction.
Cloaked in clathrin
Inside cells, clathrin, a three-legged protein, assembles many copies of itself into round cages to carry other proteins and fats. The clathrin shells shuttle their cargo from the membrane of one compartment to another. Upon delivery, the clathrin crates come apart. Pathogens sometimes hijack clathrin cages to invade and infect cells.
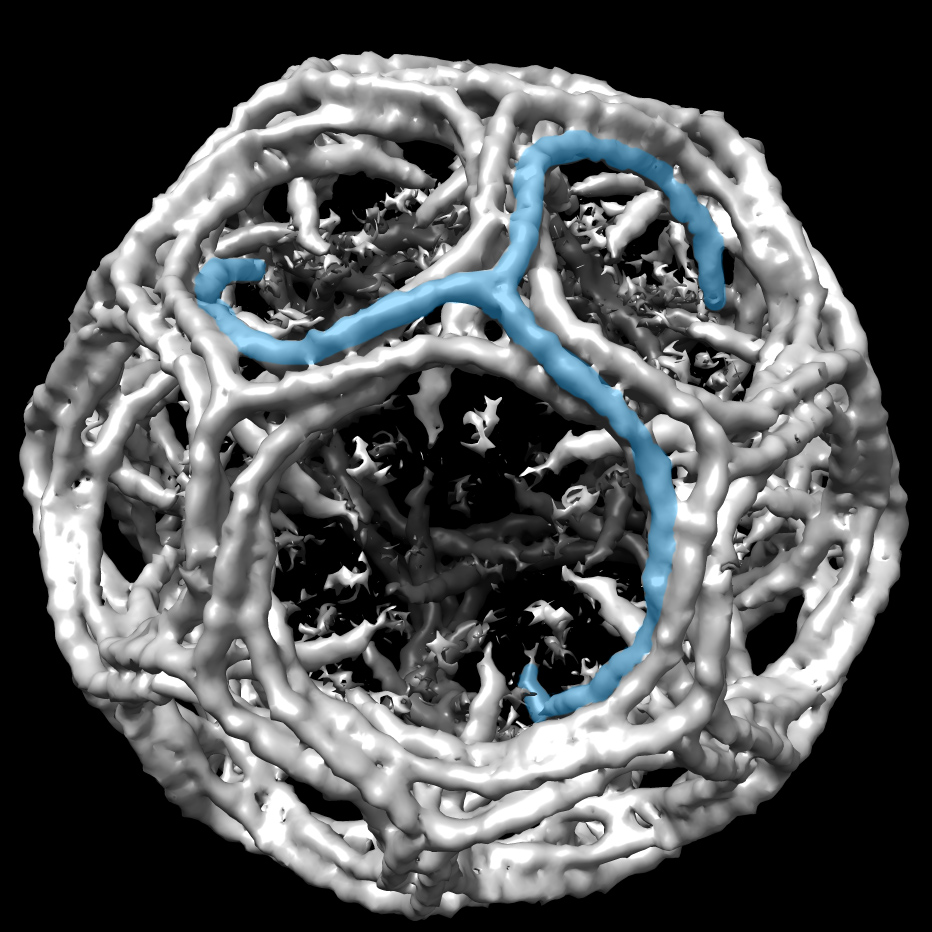
A clathrin cage with a single triskelion highlighted in blue. CryoEM map EMD_5119 was rendered in UCSF Chimera and one clathrin triskelion was highlighted. Image courtesy of Mazuraan CC BY-SA 4.0 license.
When the clathrin structure project started in the late 1990s, EM was still nicknamed “blobology” for the blurry images that seemed devoid of detailed information, points out SBGrid director Piotr Sliz, based at HMS. But those blobs so clearly represented the future of structural biology that HMS had started an EM program.
“EM was becoming more and more powerful in terms of resolution and the complexity of the structures we could analyze,” recalls Alex Fotin, who led the collaborative project as a graduate student in the HMS laboratories of Walz, Harrison and Kirchhausen. Fotin drew on the expertise of his colleagues using a mash-up of X-ray crystallography and EM.
Fotin and his colleagues analyzed two kinds of clathrin cages, a barrel and a soccer ball. “We had two pieces of information,” Fotin says. “We had the overall structure at 7.9 angstroms [using EM] and X-ray structures of small fragments at 2.6 angstrom. We were able to put the X-ray fragments into the EM map and reconstruct the structure at nearly atomic resolution.”
The team published the molecular model December 2004 in Nature. In a companion paper, the team reported the structure of clathrin with auxilin, a chaperone molecule that initiates disassembly of the clathrin cages.Fotin retired from science soon after to apply his training to the management of financial investments. “Conceptually, they use the same techniques,” says Fotin of applying machine learning, clustering, and classifying to unstructured data in market forces instead of molecular interactions.
Iron clad
Meanwhile, some of the clathrin co-authors were applying the same combination of techniques for a closer look at how iron is delivered into cells. At the time, their February 2004 Cell paper was the most finely detailed picture yet of iron and its receptor at 7.5 angstroms. The newly discovered structure of the receptor-transferrin complex reset the mechanism of iron release.
The human body has a love-hate relationship with iron. Iron is an essential nutrient, mostly for the fresh hemoglobin in the 200 billion new red blood cells made every day. Dividing cells also need iron to synthesize DNA, including cancer cells. Yet, too much of the highly reactive metal, such as in iron overload diseases, can wreak havoc by making toxic molecules that damage lipid membranes, proteins, and DNA.
This team was led by Yifan Cheng, a research associate in the HMS laboratory of EM scientist Tom Walz. “It was a great accomplishment at the time,” says Walz, who is now at The Rockefeller University in New York. Walz calls this early era “pseudoatomic resolution.” True atomic resolution in EM arrived with direct electron detectors in the last five years or so, he says.
Certain molecules were too large or flexible to crystalize. The only way to get to higher resolution was to combine intermediate resolution of single particle averaging in EM with high resolution studies from X-ray crystallography and NMR, Walz says. Single particle EM could outline the molecular envelope with bumps and bulges, while X-ray crystallography of smaller components could fill in some atomic details, such as helices, beta sheets, and amino acid side chains.
Cheng soon started his own lab at University of California, San Francisco. In 2013, he and his collaborators showed that new cryo-EM technologies were sufficient for atomic resolution of membrane proteins. They published the pain-sensing ion channel TRPV1 in open and closed formations with its chemical partners at 3.4 angstroms.
“This basically fulfilled the promise of single particle EM,” Walz says “The fact that you can get an atomic structure without making crystals with a membrane protein that was not very big made everyone realize the power of this method.”
The two early teams working with clathrin and transferrin used dedicated EM software developed by Nikolaus Grigorieff, then at nearby Brandeis University.
“It was a bit of a wild time—a gold rush,” says Grigorieff. “Many other reconstructions claimed similar resolution. People were trying new things, and it also wasn’t so clear where the dangers were. These stood the test of time.”
Grigorieff had begun developing FREALIGN as a postdoctoral fellow in the Cambridge, UK, lab of Richard Henderson, one of the 2017 Nobel Laureates, to build his own lab focusing on the single particle technique. FREALIGN is now part of a more user-friendly image processing system called CIStem, released in 2017.
Back to the Future
The third project that launched SBGrid into the EM era was a different mix of crystallography and EM techniques known as electron crystallography.
This case involved aquaporin-0, an abundant membrane protein in the eye lens of animals. Not much was known about its role, except that it looked like a specialized protein channel that conducts water molecules and also might function as molecular glue that holds the lens cells together.
The method had produced atomic resolution for others, but postdoctoral fellow Tamir Gonen in the Walz lab achieved the highest resolution structure in cryo-EM at the time at 1.9 angstrom.
Success was not assured. His aquaporin crystals were two-dimensional and thus not suitable for X-ray crystallography. Imaging the crystals in EM was also difficult, because the electron beam used to do this nudges the molecules just enough to blur the image. (Now, thanks to some advances pioneered by Grigorieff, detectors take “movies,” and software corrects for the unavoidable motion to boost resolution.)
“We abandoned imaging completely,” says Gonen, who now heads a lab at University of California, Los Angeles. Instead, he used the electron beam to produce diffraction data, which is not affected by motion, as if in X-ray crystallography. “The crystals were 2D,” Gonen says. “One pattern per crystal was collected and data from hundreds of crystals merged.” Since then, Gonen has specialized in a technique called MicroED that uses three-dimensional crystals of proteins and can evaluate small molecules.
Fast forward to 2019, cryoEM has become a standard technique used by most of the 300+ SBGrid laboratories, with about half of SBGrid effort now dedicated to maintaining cryoEM software. The work of our early pioneers revisited here was instrumental in accelerating SBGrid efforts in this area.
-Carol Cruzan Morton